Share this post
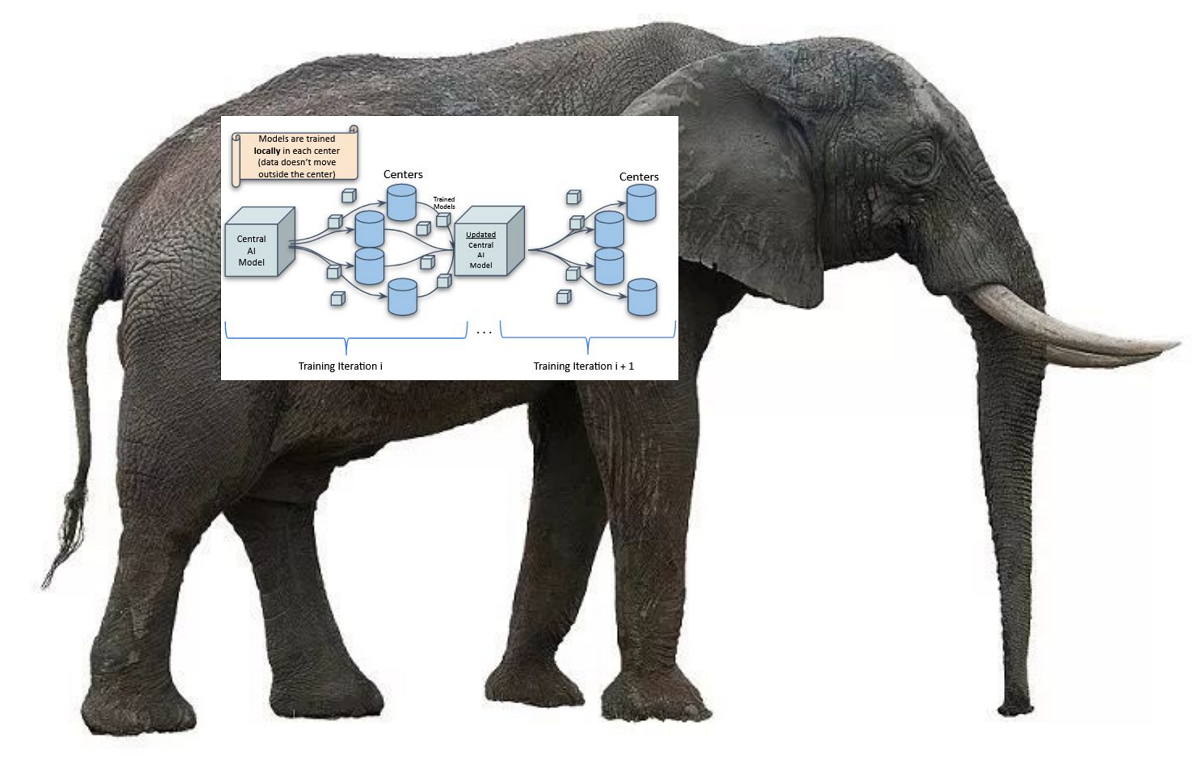
Setting up a federated network across clinical centers is like trying to eat an elephant. Yes, odd metaphor maybe, but it’s the closest one I could think of. There are ethical committees to address, institutes and hospitals to coordinate, heterogeneity in data and systems to overcome, clinical requirements to think about. I figure the list goes on.
So, first off, who am I? My name’s Akis Linardos [^1] and I just finished my first year of PhD studies in the University of Barcelona. . I’m leading a large-scale federated project, bringing clinical centers together to train a state-of-the-art breast cancer classifier. Toward this goal, we’re leveraging Flower to construct a federated network where the central server is hosted in Barcelona Supercomputing Center (BSC). So how do we go about dealing with such a massive project? Let’s take it one step at a time.
Premise 1: Communication
One rainy November night—yes, I’m a night owl—I was tapping at my keyboard, juggling multiple tabs on my browser. Delving into the source code of multiple federated frameworks, I found that what is trivial in machine learning becomes exponentially more complex in a federated learning setting. How do you pass weights around hospitals? What’s this about serializing and de-serializing my data? Where does this design choice stem from? How do I even set up the connection without running into a bloody firewall?
Well, I’m a PhD student, I should solve these issues on my own, scale the mountain and prove my intellectual prowess! Right? Wrong. The most important decision that I took was to reach out. Reach out to people that have already solved these problems, or even better—people that have developed the code and are eager to find where the users struggle. And so I came in contact with the Flower developers and now we’re on the same team quickly improving on both sides. I get to learn from them and they learn from me and the field benefits for it! Win-win.
When you’re using entirely new frameworks, it’s tempting to say, “Oh I can decrypt the inner workings of this magic box and I shouldn’t bother the developers with questions but climb this mountain myself and show the world how smart I am.” You’re smart, yes. Good for you. But if the information is out there. Use it. The developers will thank you for it.
This is just one side of the premise I’m getting at, which is: Communicate! Seriously, my mother always said being shy will ruin your life. And it will certainly ruin your research if you’re involved in real-world deployment. This doesn’t mean sending an occasional email to ask for data. It doesn’t mean putting up your best behavior until the meeting is done. It means connecting with everyone and asking the right question to push this bulky project forward. Make the meetings matter. Clinicians, data scientists, software developers and so on. Why not come in contact and educate each other? Everyone wins.
Premise 2: Start simple
There are many aspects to clinical research that I underestimated when I began my PhD. It’s Wild West compared to the polished Computer Vision, where Imagenet-sized datasets get passed around easy as candy on Halloween. And here’s why something that I learned early in my career becomes even more relevant in the planning of this project:
Breast cancer diagnosis is a big task and state of the art models often require bounding boxes or other expensive annotations. But when dealing with multiple centers, such ground truths are not easy to come by. Our solution? We start simple: Full Image Classification. For this phase, we obtain mammograms and we only ask categorical labels: Is there tumor at all? Is it Malignant? Benign? We classify based on three labels and, once we have preliminary results, we move on to the next step!
Luckily, simple doesn’t mean boring. Technical innovations on the federated algorithm are easy to try (some we’ve already tested in FeTS[^2], the first international federated challenge). The first international federated challenge. Our method, Center Dropout—which is really a simple sampling strategy of centers per round—was ranked 1st in one of the leaderboards, so how about we give it a shot in the wild?
Premise 3: Fail Fast
Here’s the added benefit to starting simple: We start fast. That means we fail fast. And yes, failing is part of the process. Undeniably. By failing I mean identifying roadblocks you didn’t anticipate. An institute uses a machine without internet connection to process data, how do we set up jobs there? Luckily, Flower’s client-side flexibility allows us to use it in very different settings. But there are more roadblocks. We ran into a firewall, how do we circumvent it? Data heterogeneity is immense between centers, how can we integrate, homogenize and prevent it in the next phase?
We have already defined many of these issues, and we have a working skeleton, but we’re prepared for more to come. It’s the way it is.
So here we are, standing side by side with the Flower developers to push this technology forward. And coming back to the question I implied early on. How do you eat an elephant? You distribute your cutlery around and feast with everyone else. And, of course, you do it one bite at a time.
Disclaimer: No wild animals were or will be hurt in the duration of this project. Especially not elephants. Did you know they are peace-makers? Yeah, they break up fights in the wild, look it up. Also, here’s a video of an elephant painting herself.
Acknowledgments: This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 952103.
[^1]: Akis Linardos: GitHub / personal website.
Share this post
