Use Partitioners¶
Understand Partitioners interactions with FederatedDataset.
Install¶
[ ]:
! pip install -q "flwr-datasets[vision]"
What is Partitioner?¶
Partitioner is an object responsible for dividing a dataset according to a chosen strategy. There are many Partitioners that you can use (see the full list here) and all of them inherit from the Partitioner object which is an abstract class providing basic structure and methods that need to be implemented for any new Partitioner to integrate with the rest of Flower Datasets code. The creation of
different Partitioner differs, but the behavior is the same = they produce the same type of objects.
IidPartitioner Creation¶
Let’s create (instantiate) the most basic partitioner, IidPartitioner and learn how it interacts with FederatedDataset.
[ ]:
from flwr_datasets.partitioner import IidPartitioner
# Set the partitioner to create 10 partitions
partitioner = IidPartitioner(num_partitions=10)
Right now the partitioner does not have access to any data therefore it has nothing to partition. FederatedDataset is responsible for assigning data to a partitioner(s).
What part of the data is assigned to partitioner?
In centralized (traditional) ML, there exist a strong concept of the splits of the dataset. Typically you can hear about train/valid/test splits. In FL research, if we don’t have an already divided datasets (e.g. by user_id), we simulate such division using a centralized dataset. The goal of that operation is to simulate an FL scenario where the data is spread across clients. In Flower Datasets you decide what split of the dataset will be partitioned. You can also resplit the datasets such
that you use a more non-custom split, or merge the whole train and test split into a single dataset. That’s not a part of this tutorial (if you are curious how to do that see Divider docs, Merger docs and preprocessor parameter docs of
FederatedDataset).
Let’s see how you specify the split for partitioning.
How do you specify the split to partition?¶
The specification of the split happens as you specify the partitioners argument for FederatedDataset. It maps partition_id: str to the partitioner that will be used for that split of the data. In the example below we’re using the train split of the cifar10 dataset to partition.
If you’re unsure why/how we chose the name of the
datasetand how to customize it, see the first tutorial.
[ ]:
from flwr_datasets import FederatedDataset
# Create the federated dataset passing the partitioner
fds = FederatedDataset(dataset="uoft-cs/cifar10", partitioners={"train": partitioner})
# Load the first partition
iid_partition = fds.load_partition(partition_id=0)
iid_partition
Dataset({
features: ['img', 'label'],
num_rows: 5000
})
[ ]:
# Let's take a look at the first three samples
iid_partition[:3]
{'img': [<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>,
<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>,
<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>],
'label': [1, 2, 6]}
Use Different Partitioners¶
Why would you need to use different ``Partitioner``s?
There are a few ways that the data partitioning is simulated in the literature. Flower Datasets let’s you work with many different approaches that have been proposed so far. It enables you to simulate partitions with different properties and different levels of heterogeneity and use those settings to evaluate your Federated Learning algorithms.
How to use different ``Partitioner``s?
To use a different Partitioner you just need to create a different object (note it has typically different parameters that you need to specify). Then you pass it as before to the FederatedDataset.
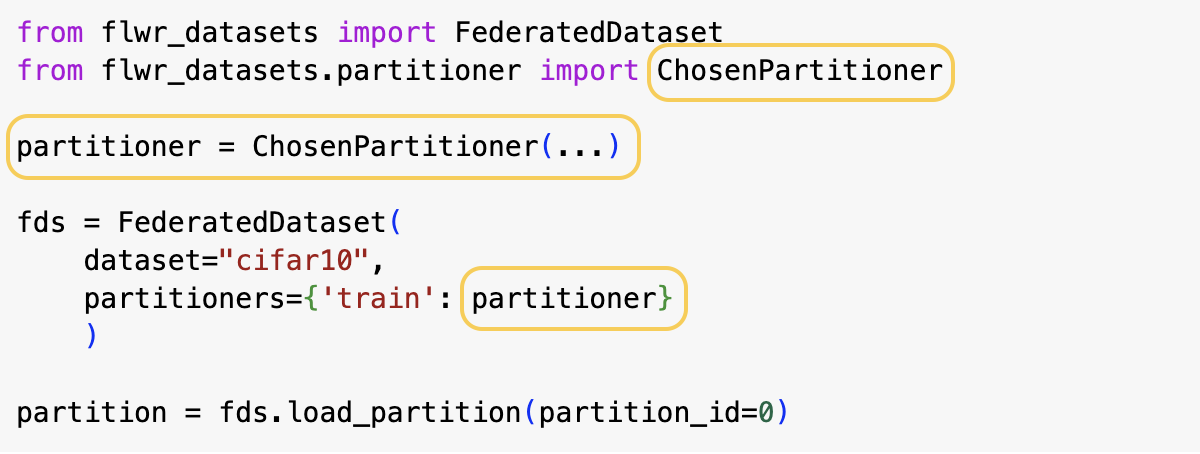
See the only changing part in yellow.
Creating non-IID partitions: Use PathologicalPartitioner¶
Now, we are going to create partitions that have only a subset of labels in each partition by using PathologicalPartitioner. In this scenario we have the exact control about the number of unique labels on each partition. The smaller the number is the more heterogeneous the division gets. Let’s have a look at how it works with
num_classes_per_partition=2.
[ ]:
from flwr_datasets.partitioner import PathologicalPartitioner
# Set the partitioner to create 10 partitions with 2 classes per partition
# Partition using column "label" (a column in the huggingface representation of CIFAR-10)
pathological_partitioner = PathologicalPartitioner(
num_partitions=10, partition_by="label", num_classes_per_partition=2
)
# Create the federated dataset passing the partitioner
fds = FederatedDataset(
dataset="uoft-cs/cifar10", partitioners={"train": pathological_partitioner}
)
# Load the first partition
partition_pathological = fds.load_partition(partition_id=0)
partition_pathological
Dataset({
features: ['img', 'label'],
num_rows: 2501
})
[ ]:
# Let's take a look at the first three samples
partition_pathological[:3]
{'img': [<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>,
<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>,
<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>],
'label': [0, 0, 7]}
[ ]:
import numpy as np
# We can use `np.unique` to get a list of the unique labels that are present
# in this data partition. As expected, there are just two labels. This means
# that this partition has only images with numbers 0 and 7.
np.unique(partition_pathological["label"])
array([0, 7])
Creating non-IID partitions: Use DirichletPartitioner¶
With the DirichletPartitioner, the primary tool for controlling heterogeneity is the alpha parameter; the smaller the value gets, the more heterogeneous the federated datasets are. Instead of choosing the exact number of classes on each partition, here we sample the probability distribution from the Dirichlet distribution, which tells how the
samples associated with each class will be divided.
[ ]:
from flwr_datasets.partitioner import DirichletPartitioner
# Set the partitioner to create 10 partitions with alpha 0.1 (so fairly non-IID)
# Partition using column "label" (a column in the huggingface representation of CIFAR-10)
dirichlet_partitioner = DirichletPartitioner(
num_partitions=10, alpha=0.1, partition_by="label"
)
# Create the federated dataset passing the partitioner
fds = FederatedDataset(
dataset="uoft-cs/cifar10", partitioners={"train": dirichlet_partitioner}
)
# Load the first partition
partition_from_dirichlet = fds.load_partition(partition_id=0)
partition_from_dirichlet
Dataset({
features: ['img', 'label'],
num_rows: 5433
})
[ ]:
# Let's take a look at the first five samples
partition_from_dirichlet[:5]
{'img': [<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>,
<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>,
<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>,
<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>,
<PIL.PngImagePlugin.PngImageFile image mode=RGB size=32x32>],
'label': [4, 4, 0, 1, 4]}
Final remarks¶
Congratulations, you now know how to use different Partitioners with FederatedDataset in Flower Datasets.
Next Steps¶
This is the second quickstart tutorial from the Flower Datasets series. See next tutorials:
Previous tutorials: